How an early variant got ahead
Throughout the COVID-19 pandemic, epidemiologists have monitored the evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with particular focus on the spike protein. An early variant with an aspartic acid (D) to glycine (G) mutation at position 614, D614G, rapidly became dominant and is maintained in current variants of concern. Zhang et al. investigated the structural basis for the increased spread of this variant, which does so even though it binds less tightly to the host receptor (see the Perspective by Choe and Farzan). Structural and biochemical studies on a full-length G614 spike trimer showed that there are interactions not present in D614 that prevent premature loss of the S1 subunit that binds angiotensin-converting enzyme 2. This stabilization effectively increases the number of spikes that can facilitate infection.
Abstract
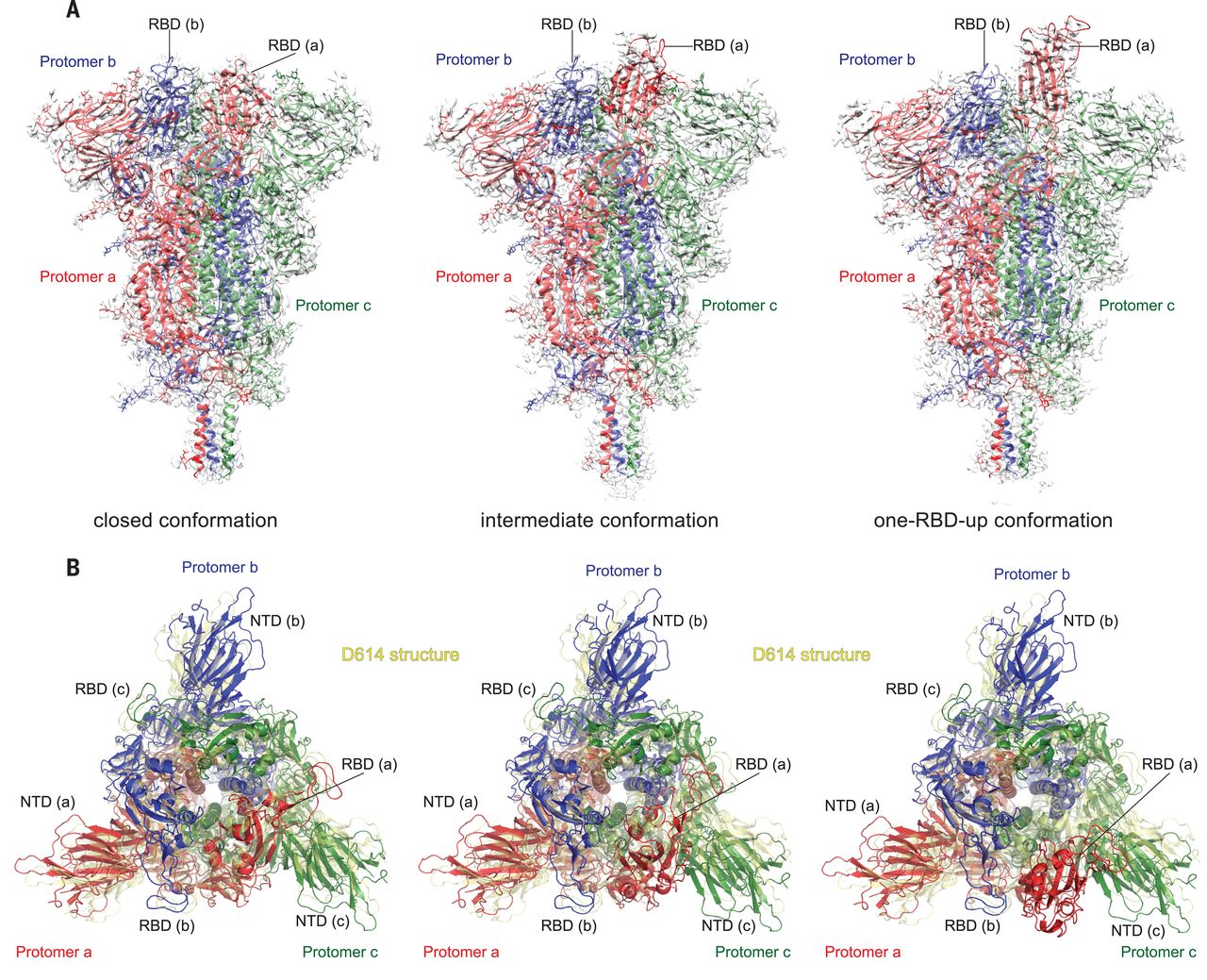
Substitution for aspartic acid (D) by glycine (G) at position 614 in the spike (S) protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) appears to facilitate rapid viral spread. The G614 strain and its recent variants are now the dominant circulating forms. Here, we report cryo–electron microscopy structures of a full-length G614 S trimer, which adopts three distinct prefusion conformations that differ primarily by the position of one receptor-binding domain. A loop disordered in the D614 S trimer wedges between domains within a protomer in the G614 spike. This added interaction appears to prevent premature dissociation of the G614 trimer—effectively increasing the number of functional spikes and enhancing infectivity—and to modulate structural rearrangements for membrane fusion. These findings extend our understanding of viral entry and suggest an improved immunogen for vaccine development.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an enveloped positive-stranded RNA virus, is the cause of the COVID-19 pandemic (1). Although the viral evolution is slowed by the RNA proofreading capability of its replication machinery (2), a variant with a single-residue substitution (D614G) in its spike (S) protein rapidly became the dominant strain throughout the world (3). It has since further evolved to give several variants of concern (VOCs) (4–6). The trimeric S protein decorates the viral surface and is an important target for the development of diagnostics, therapeutics, and vaccines; therefore, understanding the effect of key mutations may guide intervention strategies. Here, we focus on the D614G mutation that is in all currently circulating strains of SARS-CoV-2. The S protein is produced as a single-chain precursor and is subsequently processed by a furin-like protease into the receptor-binding fragment S1 and the fusion fragment S2 (7). After engagement of the receptor-binding domain (RBD) in S1 with the viral receptor angiotensin-converting enzyme 2 (ACE2) on the host cell surface, followed by a second proteolytic cleavage within S2 (S2′ site) (8), the S protein undergoes large conformational changes, which result in the dissociation of S1 and the irreversible refolding of S2 into a postfusion structure (9, 10). This induces fusion of the virus and host cell membranes to initiate infection. Rapid advances in structural biology of the SARS-CoV-2 S protein include structures of its soluble fragments: the ectodomain stabilized in its prefusion conformation (11–13), RBD-ACE2 complexes (14–17), and segments of S2 in the postfusion state (18). In the prefusion ectodomain structure, S1 folds into four domains—the N-terminal domain (NTD), the RBD, and two C-terminal domains (CTDs)—and wraps around the prefusion S2, with the RBD sampling two distinct conformations: up for a receptor-accessible state and down for a receptor-inaccessible state. We and others have reported structures of a purified, full-length D614 S protein in both prefusion and postfusion conformations (19, 20). Studies by cryo–electron tomography (cryo-ET), with chemically inactivated SARS-CoV-2 preparations, using both D614 and G614 variants have revealed additional structural details of S proteins present on the surface of the virion (21–24).
Epidemiological surveillance has indicated that the SARS-CoV-2 carrying G614 outcompeted the original virus and became the globally dominant form within a month (3, 25, 26). This single-residue substitution appears to correlate with high viral loads in infected patients and high infectivity of pseudotyped viruses, but not with disease severity (3). The G614 virus has comparable sensitivity to neutralization by convalescent human sera or vaccinated hamster sera (3, 27–30), which suggests that vaccines containing D614 remain effective against the G614 virus. Moreover, S1 dissociates more readily from the D614 virus than from G614 virus (31), which indicates that the D614 viral spike is less stable than that of the G614 variant. The G614 ectodomain trimer is reported to sample the RBD-up conformations more frequently than the D614 trimer (13, 29, 32), but it is puzzling why the former binds more weakly to recombinant ACE2 than the latter (32). The known S trimer structures indicate that the D614G change breaks a salt bridge between D614 and a lysine residue (K854) in the fusion peptide–proximal region (FPPR) (19, 33, 34), which may help clamp the RBD in the prefusion conformation. This observation can explain why the G614 trimer favors the RBD-up conformations but does not account for its increased stability. To resolve these issues, we report the structural consequences of the D614G substitution in the context of the full-length S protein.
We compared the membrane fusion activity of the full-length G614 S protein (fig. S1) with that of the D614 S construct in a cell-cell fusion assay (19). All of the cells expressing S fused efficiently with cells transfected with a human ACE2 construct (fig. S2A), demonstrating that the S proteins expressed on the cell surfaces are fully functional. At low transfection levels, G614 S had higher fusion activity than the D614 S, but the difference diminished with the increased amount of transfected DNA, which suggests that the high expression levels can compensate for lower fusion efficiency of the D614 S protein. The G614 trimer remains sensitive to inhibition by an engineered trimeric ACE2-based inhibitor that competes with the receptor on the target cells (35) (fig. S2B). For protein purification, we used a construct fused with a C-terminal strep-tag, which was equally active in cell-cell fusion as the untagged version (fig. S2A), and purified both G614 and D614 proteins under identical conditions. The D614 protein eluted in three peaks characterized previously as the prefusion S trimer, the postfusion S2 trimer, and the dissociated monomeric S1 (19). The G614 protein eluted as a single major peak, corresponding to the prefusion trimer (Fig. 1A). This suggests that D614G has a notable effect on the stability of the SARS-CoV-2 S trimer. Coomassie-stained SDS–polyacrylamide gel electrophoresis (SDS-PAGE) analysis shows that G614 elutes mainly as the prefusion trimer, comprising the cleaved S1-S2 complex (~90%) and a small amount of the uncleaved S precursor (~10%). We next measured binding of the prefusion trimer fractions of the full-length proteins to recombinant soluble ACE2 by biolayer interferometry (BLI) (Fig. 1B). The S trimers bound more strongly to a dimeric ACE2 than to a monomeric ACE2, as expected. The G614 protein bound ACE2 less tightly than the D614 protein, which is consistent with the measurements reported by others using soluble constructs (32). This observation appears inconsistent with accounts that the G614 trimer has a more exposed RBD than the D614 trimer (13, 21, 22, 29, 32). We note that the second binding event between dimeric ACE2 and a G614 trimer has both a slower on-rate and a slower off-rate than that for a D614 trimer (table S1).
Full article on: